Calpain (EC 3.4.22.17, Clan CA, family C02) has a substrate specificity that is relatively restricted, and most of oligopeptides are not efficiently hydrolyzed. Casein is the most popular substrate for in vitro assays, and it is used either as the natural protein or modified with various chromophores, fluorescent reagents or isotopes. Calpain purified from skeletal muscle by standard methods has a specific activity of several hundred units per mg protein, where 1 unit corresponds to an increase of 1.0 absorbance unit at 280 nm per hour under the standard assay conditions (if 1 unit of calpain is incubated in 0.5 ml of 3 mg/ml casein, 0.1 M Tris-HCl (pH 7.5), 25 mM of 2-mercaptoethanol, and appropriate concentrations of CaCl2 at 30oC for 20 min, the acid-soluble supernatant made by adding 0.5 ml of 10% trichloroacetic acid and centrifugation shows an increase of 0.333 absorbance unit at 280 nm). Activity is dependent on the Ca2+ concentration, and that giving half maximal activity for μ- and m-calpains are around 50 μM and 0.3 mM, respectively.
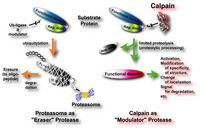
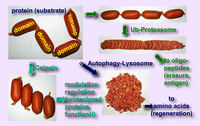
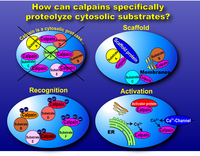
The rules governing the specificity of the calpains remain unclear. This is the main motif for this Calpain for Modulatory Proteolysis Database (CaMPDB) project. It seems that calpain recognizes the overall 3-dimensional structure of its substrates more than the primary structure. Even so, hydrophobic (Tyr, Met, Leu, Val) and Arg residues tend to be preferred in position P2 (Brown & Crawford 1993; Stabach et al. 1997; Tompa et al. 2004). Protein kinases, phosphatases, phospholipases, cytoskeletal proteins, membrane proteins, cytokines, transcription factors, lens proteins, calmodulin-binding proteins and others have been suggested to be in vivo substrates, but clear evidence has not yet been obtained. Calpain proteolyzes these proteins in a limited manner rather than digesting them to small peptides, indicating its modulatory functions for the substrate proteins by cutting their interdomain regions (Saido et al. 1994b; Sorimachi et al. 1997) (Fig. 1-3). In the following sections, several representative examples are to be described.
The activity and specificity of m-calpain are almost identical to those of μ-calpain except for the Ca2+ requirement. Calpain has a very specific in vivo protein inhibitor, named calpastatin. Calpastatin contains four repeats of the inhibitory unit, each of which can inhibit calpain independently. Both μ- and m-calpains have similar susceptibility to calpastatin. Most inhibitors, including calpastatin, that are efficient for μ-calpain are efficient for m-calpain with few exceptions (Li et al. 1996).
The Ca2+ concentration required for activation of μ-calpain (pKa) is lowered by the addition of phosphoinositides such as PIP, PIP2, PIP3 (Saido et al. 1992). In the presence of phosphoinositides, the Ca2+ concentration required for the activity is reduced to the range 100nM-1μM. Several activator proteins have now been reported (Melloni et al. 2000; Melloni et al. 1998; Tompa et al. 2002). During the activation of μ-calpain, autolysis of the N-terminal few residues occurs. This autolysis precedes the appearance of proteolytic activity in the normal conditions in vitro, and thus has been considered to be an essential part of the activation mechanism (Saido et al. 1994a). Recently, another role of autolysis has been revealed in m-calpain, i.e., it is involved in subunit dissociation (Nakagawa et al. 2001).
N-terminal autolysis of m-calpain has long been believed to be an activation process. However, a recent study indicates that autolysis of the N-terminus is not required for activation, but is involved in the association and dissociation of the two subunits (Nakagawa et al. 2001). Lys7 and Arg12 of the N-terminus of mCL make salt-bridges with Asp154 and Glu260 of EF-2 and EF-5 of the 30K/CAPNS1, which is identical to the regulatory subunit of μ-calpain. The amino acid sequences of human mCL and μCL are 63% identical, and have the same domain structure (see “Calpain Overview” Fig. 2).
2. Substrates of calpains in brain function
Calpain is believed to be strongly related to certain brain functions; the involvement of calpain in ischemia was first suggested in 1989 (Kuwaki et al. 1989). Since then, there have been many reports that examined the relationship between calpain and ischemia (Kampfl et al. 1997; Saido et al. 1994b; Siman et al. 1996). For example, drastic spatial changes were observed in the proteolysis of fodrin in the hippocampus after 1-24 hours of transient ischemia using a proteolyzed α-fodrin 150 kDa-fragment-specific antibody. Fodrin is a major cytoskeletal protein and one of the best substrates for calpain. The method developed in these studies is widely applicable, and has been used not only in studies of ischemia but also in various other proteinase studies such as β-amyloid processing, integrin proteolysis, and calpain autolysis. Calpain inhibitors can suppress these post-ischemic changes as described below, suggesting a therapeutic potential for calpain inhibitors in brain ischemia. These observations suggest that calpain functions as a mediator of the pathological process, not necessarily as the ultimate destroyer of cells.
Calpain may also be involved in long-term potentiation and Alzheimer’s disease. m-calpain was reported to be over-expressed in the brain of patients with Alzheimer’s disease (Grynspan et al. 1997). An interesting question that needs explanation is why ubiquitously expressed μ- and/or m-calpains are specifically involved in brain-specific functions.
The inhibitors described above effectively prevent fodrinolysis upon ischemia, and could be applied as therapeutic agents in the future. For example, AK295 was shown to attenuate motor and cognitive deficits following experimental brain injury in rats (Saatman et al. 1996). Thus, calpain inhibitors are one of the most competitive molecules for development in the near future. The road to success is, of course, the specificity of the inhibitor.
The p53 protein plays an essential role in tumor suppression, and mutational defects in p53 function are the most frequently detected genetic event in human cancers. Most normal cells express very low levels of p53 due to the relatively short half-life of the protein. There is evidence that p53 can be degraded through ubiquitin-dependent proteolysis. p53 interacts with the E6 protein encoded by the human papilloma virus, and E6 in association with the cell protein E6-AP can function as a ubiquitin ligase that targets p53 for degradation. On the other hand, several groups independently reported that p53 is sensitively proteolyzed, at least in vitro, and may be regulated by calpain (Gonen et al. 1997; Kubbutat & Vousden 1997; Pariat et al. 1997; Zhang et al. 1997). Kubbutat and Vousden showed that calpain cleaves p53 in vitro, and that calpain inhibitors I (16 μM in the medium) and II (100 to 200 μM) enhance endogenous p53 levels in MCF-7 (breast carcinoma) and RKO (colon cancer) cells. They also showed that the E6/ubiquitin-dependent degradation of p53 is not affected by calpain inhibitor I (up to 200 μM). Pariat et al. also showed that oligomerization of p53 through the C-terminal domain does not affect the cleavage kinetics, but that heat denatured p53, which retains its sensitivity to trypsin, is resistant to calpain proteolysis. Their results indicate that an intact p53 quaternary structure is not a prerequisite for cleavage, but that the tertiary structure is essential for recognition by calpain. Furthermore, they observed that the over-expression of calpastatin in H358a and SAOS cells leads to the accumulation of p53 in a dose-dependent manner, and that the addition of the Ca2+ ionophore A21187 reduces p53 levels, an effect that can be prevented by the further addition of E-64d, calpain inhibitor I, or calpain inhibitor II. Ciechanover’s group also showed in in vitro experiments that calpain can digest p53 as well as N-myc, c-Fos, or c-Jun, and that the digestion can be stopped by E-64, EGTA, MG115 (carbobenzoxyl-leucinyl-leucinyl-norvalinal), or calpain inhibitor I or II (Gonen et al. 1997). Mellgren’s group used a calpain-selective cell permeable inhibitor, benzyloxycarbonyl Leu-Leu-Tyr diazomethylketone, on serum-stimulated WI-38 human fibroblasts, and observed similar results (Zhang et al. 1997).
In addition, the susceptibility of naturally occurring p53 mutants to calpain was examined. These mutants can be classified into 3 groups based on their susceptibility to calpain. The first group includes Ser15Ala, Ser15Asp, Met246Val, Arg256Ala, Val272Met, and Arg273Cys, all showing sensitivities similar to that of wild type p53. The second group, including Ala138Val, Arg175His, Met237Ile, and Arg273Pro, show reduced sensitivity, while the third group, including Arg248Trp, Arg273Cys, and Arg273His, are more sensitive to calpain. These results strongly suggest the importance of the local tertiary structure of p53 in its proteolysis by calpain.
These results described above are essentially in vitro experiments. To prove the involvement of calpain in cellular functions, it is important to obtain more rigorous evidence that it is actually involved in in vivo regulation of p53. However, unlike proteasome, the ability of calpain to process substrates into only a few fragments is suitable for modulating such ubiquitous and functional proteins as p53. One critical question is whether calpain truly functions in the nucleus in vivo. In this regard, Kubbutat and Vousden (Kubbutat & Vousden 1997) observed calpain activity in the nuclear fraction, and Mellgren’s group (Zhang et al. 1997) reported that μ-calpain, but not m-calpain, is transported into nuclei in an ATP-dependent fashion. Thus, it is possible that calpain translocates into the nucleus under certain cell conditions, resulting in interaction with and regulation of the p53 molecule. It should be noted that oncoprotein Mdm2 was reported to promote the rapid degradation of p53, and this pathway is under the regulation of proteasome, not calpain.
Apoptosis is the common phenotype of programmed or physiologic cell death, a process in which caspases play important roles. The possible involvement of calpain in apoptosis was first suggested in 1993 (Roberts-Lewis et al. 1993; Sarin et al. 1993), and has been reported for various cells including thymocytes, hippocampal neurons, and hepatocytes. Two of the hallmarks of apoptosis are the proteolytic cleavage of fodrin and DNA fragmentation. As previously reported, fodrin is a very good in vitro substrate for calpain, and is thought to be one of the physiological substrates. Thus, it is suspected that calpain is involved in apoptosis via fodrinolysis, but fodrinolysis coupling with apoptosis does not seem to be related to calpain. In either case, observations concerning apoptosis differ depending on the cell line studies, and it is not yet established whether the fundamental mechanism of apoptosis is identical for all cells. Within a given cell, however, it seems that various distinct triggers can produce a very similar final apoptotic phenotype. In thymocytes, for example, Squier and Cohen (Squier & Cohen 1997) recently observed that PD150606 (ca. 10 μM) as well as E-64d (300 μM), MDL 28170 (carbenzoxy-Val-phenylalaninal), and calpain inhibitor I (20 μM) prevent dexamethasone-induced apoptosis in thymocytes, but that valinomycin- and heat shock-induced apoptosis are not inhibited. Moreover, DNA fragmentation induced by the addition of Ca2+ (5 mM in medium) is not inhibited by calpain inhibitor I (20 μM). Based on these results, they argued that calpain acts as a factor in the initiation of apoptosis of thymocytes, rather than in a final common pathway or mechanism. Thus, the involvement of calpain seems to be independent of the caspase family, and the interaction between p53 and calpain described above is not unrelated to calpain-mediated apoptosis, since p53 also triggers apoptosis.
In addition, a number of calpain homologues in Caenorhabditis elegans may be related to the well-studied apoptotic mechanisms in nematodes involving the ced-3 gene product. It should be noted, however, that the specificity of the inhibitors used for experiments is crucial in interpreting these results, and only the specific inhibition of calpain can lead to a correct interpretation of the experimental results. The effects of proteasome and lysozomal proteases must be eliminated in order to obtain a true understanding of the mechanism of calpain-mediated apoptosis.
- Arthur, J.S., Elce, J.S., Hegadorn, C., Williams, K. & Greer, P.A. (2000) Disruption of the murine calpain small subunit gene, Capn4: calpain is essential for embryonic development but not for cell growth and division. Mol. Cell Biol. 20, 4474-4481.
- Azam, M., Andrabi, S.S., Sahr, K.E., Kamath, L., Kuliopulos, A. & Chishti, A.H. (2001) Disruption of the mouse mu-calpain gene reveals an essential role in platelet function. Mol. Cell. Biol. 21, 2213-2220.
- Barnes, T.M. & Hodgkin, J. (1996) The tra-3 sex determination gene of Caenorhabditis elegans encodes a member of the calpain regulatory protease family. EMBO J. 15, 4477-4484.
- Brown, N. & Crawford, C. (1993) Structural modifications associated with the change in Ca2+ sensitivity on activation of m-calpain. FEBS Lett 322, 65-68.
- Cao, Y., Zhao, H. & Grunz, H. (2001) XCL-2 is a novel m-type calpain and disrupts morphogenetic movements during embryogenesis in Xenopus laevis. Dev. Growth Differ. 43, 563-571.
- Dayton, W.R., Goll, D.E., Zeece, M.G., Robson, R.M. & Reville, W.J. (1976) A Ca2+-activated protease possibly involved in myofibrillar protein turnover. Purification from porcine muscle. Biochemistry 15, 2150-2158.
- Dear, N., Matena, K., Vingron, M. & Boehm, T. (1997) A new subfamily of vertebrate calpains lacking a calmodulin-like domain: implications for calpain regulation and evolution. Genomics 45, 175-184.
- Delaney, S.J., Hayward, D.C., Barleben, F., Fischbach, K.F. & Miklos, G.L. (1991) Molecular cloning and analysis of small optic lobes, a structural brain gene of Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 88, 7214-7218.
- Denison, S.H., Orejas, M. & Arst, H.N., Jr. (1995) Signaling of ambient pH in Aspergillus involves a cysteine protease. J. Biol. Chem. 270, 28519-28522.
- Diez, E., Alvaro, J., Espeso, E.A., et al. (2002) Activation of the Aspergillus PacC zinc finger transcription factor requires two proteolytic steps. EMBO J. 21, 1350-1359.
- Drummond, G.I. & Duncan, L. (1968) On the mechanism of activation of phosphorylase b kinase by calcium. J Biol Chem 243, 5532-5538.
- Dutt, P., Croall, D.E., Arthur, S.C., et al. (2006) m-Calpain is required for preimplantation embryonic development in mice. BMC Dev. Biol. 6, DOI:10.1186/1471-1213X-1186-1183.
- Ersfeld, K., Barraclough, H. & Gull, K. (2005) Evolutionary relationships and protein domain architecture in an expanded calpain superfamily in kinetoplastid parasites. J Mol Evol 61, 742-757.
- Futai, E., Maeda, T., Sorimachi, H., Kitamoto, K., Ishiura, S. & Suzuki, K. (1999) The protease activity of a calpain-like cysteine protease in Saccharomyces cerevisiae is required for alkaline adaptation and sporulation. Mol. Gen. Genet. 260, 559-568.
- Goll, D.E., Thompson, V.F., Li, H., Wei, W. & Cong, J. (2003) The calpain system. Physiol. Rev. 83, 731-801.
- Gonen, H., Shkedy, D., Barnoy, S., Kosower, N.S. & Ciechanover, A. (1997) On the involvement of calpains in the degradation of the tumor suppressor protein p53. FEBS Lett. 406, 17-22.
- Grynspan, F., Griffin, W.B., Mohan, P.S., Shea, T.B. & Nixon, R.A. (1997) Calpains and calpastatin in SH-SY5Y neuroblastoma cells during retinoic acid-induced differentiation and neurite outgrowth: comparison with the human brain calpain system. J Neurosci Res 48, 181-191.
- Guroff, G. (1964) A neutral calcium-activated proteinase from the soluble fraction of rat brain. J. Biol. Chem. 239, 149-155.
- Hata, S., Nishi, K., Kawamoto, T., et al. (2001) Both the conserved and the unique gene structure of stomach-specific calpains reveal processes of calpain gene evolution. J Mol Evol 53, 191-203.
- Hayashi, M., Fukuzawa, T., Sorimachi, H. & Maeda, T. (2005) Constitutive activation of the pH-responsive Rim101 pathway in yeast mutants defective in late steps of the MVB/ESCRT pathway. Mol. Cell. Biol. 25, 9478-9490.
- Herranz, S., Rodriguez, J.M., Bussink, H.J., et al. (2005) Arrestin-related proteins mediate pH signaling in fungi. Proc. Natl. Acad. Sci. U.S.A. 102, 12141-12146.
- Horikawa, Y., Oda, N., Cox, N.J., et al. (2000) Genetic variation in the gene encoding calpain-10 is associated with type 2 diabetes mellitus. Nat. Genet. 26, 163-175.
- Huston, R.B. & Krebs, E.G. (1968) Activation of skeletal muscle phosphorylase kinase by Ca2+. II. Identification of the kinase activating factor as a proteolytic enzyme. Biochemistry 7, 2116-2122.
- Ishiura, S., Murofushi, H., Suzuki, K. & Imahori, K. (1978) Studies of a calcium-activated neutral protease from chicken skeletal muscle. I. Purification and characterization. J. Biochem. (Tokyo) 84, 225-230.
- Kammenga, J.E., Doroszuk, A., Riksen, J.A., et al. (2007) A Caenorhabditis elegans Wild Type Defies the Temperature-Size Rule Owing to a Single Nucleotide Polymorphism in tra-3. PLoS Genet 3, e34.
- Kampfl, A., Posmantur, R.M., Zhao, X., Schmutzhard, E., Clifton, G.L. & Hayes, R.L. (1997) Mechanisms of calpain proteolysis following traumatic brain injury: implications for pathology and therapy: implications for pathology and therapy: a review and update. J Neurotrauma 14, 121-134.
- Kubbutat, M.H. & Vousden, K.H. (1997) Proteolytic cleavage of human p53 by calpain: a potential regulator of protein stability. Mol. Cell. Biol. 17, 460-468.
- Kuwaki, T., Satoh, H., Ono, T., Shibayama, F., Yamashita, T. & Nishimura, T. (1989) Nilvadipine attenuates ischemic degradation of gerbil brain cytoskeletal proteins. Stroke 20, 78-83.
- Lee, H.J., Sorimachi, H., Jeong, S.Y., Ishiura, S. & Suzuki, K. (1998) Molecular cloning and characterization of a novel tissue-specific calpain predominantly expressed in the digestive tract. Biol. Chem. 379, 175-183.
- Lee, H.J., Tomioka, S., Kinbara, K., et al. (1999) Characterization of a human digestive tract-specific calpain, nCL-4, expressed in the baculovirus system. Arch. Biochem. Biophys. 362, 22-31.
- Li, Z., Ortega-Vilain, A.C., Patil, G.S., et al. (1996) Novel peptidyl alpha-keto amide inhibitors of calpains and other cysteine proteases. J Med Chem 39, 4089-4098.
- Lid, S.E., Gruis, D., Jung, R., et al. (2002) The defective kernel 1 (dek1) gene required for aleurone cell development in the endosperm of maize grains encodes a membrane protein of the calpain gene superfamily. Proc. Natl. Acad. Sci. U.S.A. 99, 5460-5465.
- Liu, K., Li, L. & Cohen, S.N. (2000) Antisense RNA-mediated deficiency of the calpain protease, nCL-4, in NIH3T3 cells is associated with neoplastic transformation and tumorigenesis. J Biol Chem 275, 31093-31098.
- Melloni, E., Averna, M., Salamino, F., Sparatore, B., Minafra, R. & Pontremoli, S. (2000) Acyl-CoA-binding protein is a potent m-calpain activator. J Biol Chem 275, 82-86.
- Melloni, E., Michetti, M., Salamino, F. & Pontremoli, S. (1998) Molecular and functional properties of a calpain activator protein specific for mu-isoforms. J Biol Chem 273, 12827-12831.
- Meyer, W.L., Fischer, E.H. & Krebs, E.G. (1964) Activation of skeletal muscle phosphorylase kinase by Ca2+. Biochemistry 3, 1033-1039.
- Mugita, N., Kimura, Y., Ogawa, M., Saya, H. & Nakao, M. (1997) Identification of a novel, tissue-specific calpain htra-3; a human homologue of the Caenorhabditis elegans sex determination gene. Biochem Biophys Res Commun 239, 845-850.
- Nakagawa, K., Masumoto, H., Sorimachi, H. & Suzuki, K. (2001) Dissociation of m-calpain subunits occurs after autolysis of the N-terminus of the catalytic subunit, and is not required for activation. J. Biochem. 130, 605-611.
- Ohno, S., Emori, Y., Imajoh, S., Kawasaki, H., Kisaragi, M. & Suzuki, K. (1984) Evolutionary origin of a calcium-dependent protease by fusion of genes for a thiol protease and a calcium-binding protein? Nature 312, 566-570.
- Ono, Y., Shimada, H., Sorimachi, H., et al. (1998) Functional defects of a muscle-specific calpain, p94, caused by mutations associated with limb-girdle muscular dystrophy type 2A. J. Biol. Chem. 273, 17073-17078.
- Ono, Y., Sorimachi, H. & Suzuki, K. (1999) New aspect of the research on limb-girdle muscular dystrophy 2A: a molecular biologic and biochemical approach to pathology. Trends Cardiovasc Med 9, 114-118.
- Ono, Y., Torii, F., Ojima, K., et al. (2006) Suppressed disassembly of autolyzing p94/CAPN3 by N2A connectin/titin in a genetic reporter system. J. Biol. Chem. 281, 18519-18531.
- Pariat, M., Carillo, S., Molinari, M., et al. (1997) Proteolysis by calpains: a possible contribution to degradation of p53. Mol. Cell. Biol. 17, 2806-2815.
- Penas, M.M., Hervas-Aguilar, A., Munera-Huertas, T., et al. (2007) Further characterization of the signaling proteolysis step in the Aspergillus nidulans pH signal transduction pathway. Eukaryot. Cell, in press.
- Richard, I., Broux, O., Allamand, V., et al. (1995) Mutations in the proteolytic enzyme calpain 3 cause limb-girdle muscular dystrophy type 2A. Cell 81, 27-40.
- Roberts-Lewis, J.M., Marcy, V.R., Zhao, Y., Vaught, J.L., Siman, R. & Lewis, M.E. (1993) Aurintricarboxylic acid protects hippocampal neurons from NMDA- and ischemia-induced toxicity in vivo. J. Neurochem. 61, 378-381.
- Saatman, K.E., Murai, H., Bartus, R.T., et al. (1996) Calpain inhibitor AK295 attenuates motor and cognitive deficits following experimental brain injury in the rat. Proc. Natl. Acad. Sci. U.S.A. 93, 3428-3433.
- Saido, T.C., Nagao, S., Shiramine, M., et al. (1994a) Distinct kinetics of subunit autolysis in mammalian m-calpain activation. FEBS Lett 346, 263-267.
- Saido, T.C., Shibata, M., Takenawa, T., Murofushi, H. & Suzuki, K. (1992) Positive regulation of mu-calpain action by polyphosphoinositides. J Biol Chem 267, 24585-24590.
- Saido, T.C., Sorimachi, H. & Suzuki, K. (1994b) Calpain: new perspectives in molecular diversity and physiological-pathological involvement. FASEB J. 8, 814-822.
- Sarin, A., Adams, D.H. & Henkart, P.A. (1993) Protease inhibitors selectively block T cell receptor-triggered programmed cell death in a murine T cell hybridoma and activated peripheral T cells. J. Exp. Med. 178, 1693-1700.
- Siman, R., Bozyczko-Coyne, D., Savage, M.J. & Roberts-Lewis, J.M. (1996) The calcium-activated protease calpain I and ischemia-induced neurodegeneration. Adv Neurol 71, 167-174; discussion 174-165.
- Sokol, S.B. & Kuwabara, P.E. (2000) Proteolysis in Caenorhabditis elegans sex determination: cleavage of TRA-2A by TRA-3. Genes Dev 14, 901-906.
- Sorimachi, H., Imajoh-Ohmi, S., Emori, Y., et al. (1989) Molecular cloning of a novel mammalian calcium-dependent protease distinct from both m- and mu-types. Specific expression of the mRNA in skeletal muscle. J. Biol. Chem. 264, 20106-20111.
- Sorimachi, H., Ishiura, S. & Suzuki, K. (1993a) A novel tissue-specific calpain species expressed predominantly in the stomach comprises two alternative splicing products with and without Ca2+-binding domain. J. Biol. Chem. 268, 19476-19482.
- Sorimachi, H., Ishiura, S. & Suzuki, K. (1997) Structure and physiological function of calpains. Biochem J 328 ( Pt 3), 721-732.
- Sorimachi, H. & Suzuki, K. (2001) The structure of calpain. J. Biochem. 129, 653-664.
- Sorimachi, H., Toyama-Sorimachi, N., Saido, T.C., et al. (1993b) Muscle-specific calpain, p94, is degraded by autolysis immediately after translation, resulting in disappearance from muscle. J. Biol. Chem. 268, 10593-10605.
- Sorimachi, H., Tsukahara, T., Okada-Ban, M., Sugita, H., Ishiura, S. & Suzuki, K. (1995) Identification of a third ubiquitous calpain species--chicken muscle expresses four distinct calpains. Biochim Biophys Acta 1261, 381-393.
- Squier, M.K. & Cohen, J.J. (1997) Calpain, an upstream regulator of thymocyte apoptosis. J. Immunol. 158, 3690-3697.
- Stabach, P.R., Cianci, C.D., Glantz, S.B., Zhang, Z. & Morrow, J.S. (1997) Site-directed mutagenesis of alpha II spectrin at codon 1175 modulates its mu-calpain susceptibility. Biochemistry 36, 57-65.
- Suzuki, K. (1991) Nomenclature of calcium dependent proteinase. Biomed Biochim Acta 50, 483-484.
- Suzuki, K., Hata, S., Kawabata, Y. & Sorimachi, H. (2004) Structure, activation, and biology of calpain. Diabetes 53, S12-18.
- Syntichaki, P., Xu, K., Driscoll, M. & Tavernarakis, N. (2002) Specific aspartyl and calpain proteases are required for neurodegeneration in C. elegans. Nature 419, 939-944.
- Tompa, P., Buzder-Lantos, P., Tantos, A., et al. (2004) On the sequential determinants of calpain cleavage. J Biol Chem 279, 20775-20785.
- Tompa, P., Mucsi, Z., Orosz, G. & Friedrich, P. (2002) Calpastatin subdomains A and C are activators of calpain. J Biol Chem 277, 9022-9026.
- Tonami, K., Kurihara, Y., Aburatani, H., Uchijima, Y., Asano, T. & Kurihara, H. (2007) Calpain 6 Is Involved in Microtubule Stabilization and Cytoskeletal Organization. Mol Cell Biol.
- Yoshikawa, Y., Mukai, H., Hino, F., Asada, K. & Kato, I. (2000) Isolation of two novel genes, down-regulated in gastric cancer. Jpn J Cancer Res 91, 459-463.
- Zhang, W., Lu, Q., Xie, Z.J. & Mellgren, R.L. (1997) Inhibition of the growth of WI-38 fibroblasts by benzyloxycarbonyl-Leu-Leu-Tyr diazomethyl ketone: evidence that cleavage of p53 by a calpain-like protease is necessary for G1 to S-phase transition. Oncogene 14, 255-263.
- Zimmerman, U.J., Boring, L., Pak, J.H., Mukerjee, N. & Wang, K.K. (2000) The calpain small subunit gene is essential: its inactivation results in embryonic lethality. IUBMB Life 50, 63-68.